Our Datasets
Open Medical & Sports Datasets
 1
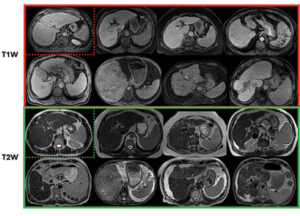
1CirrMRI600+
628 abdominal MRI volumes (T1W: 310, T2W: 318) with physician masks for liver cirrhosis research; single-center, multivendor, multisequence.
MRISegmentationLiver
 2
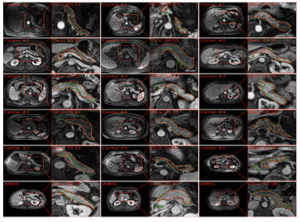
2PanSegData
767 MRI and 1,350 CT scans for pancreas segmentation. PanSegNet achieved Dice: 88.3% (CT), 85.0% (T1W), 86.3% (T2W); strong volume correlation and agreement.
MRICTSegmentation
 3
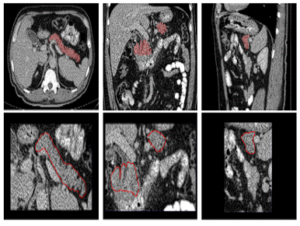
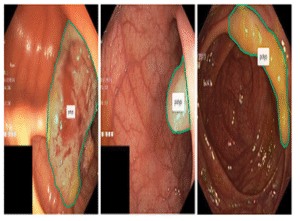
3Peri-Pancreatic Edema
255 pancreatitis CT scans with edema labels (179 positive, 76 negative) and expert pancreas masks to support robust research.
CTClassification
 4
4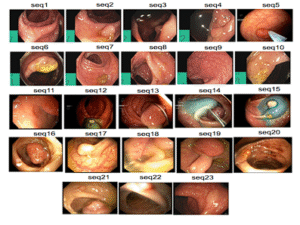 5
5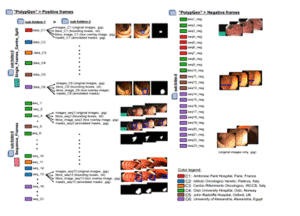 6
6 7
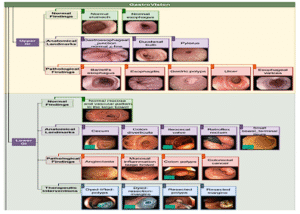
7ImageCLEFmed MEDVQA GI 2023
Colonoscopy images with text and segmentation for VQA, VQG, and VLQA tasks to enhance interpretability in diagnostics.
EndoscopyVQASegmentation
 8
8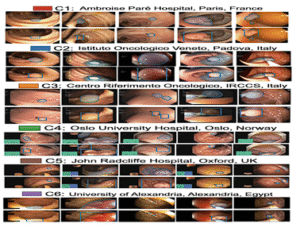 9
9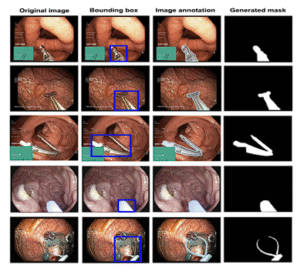 10
10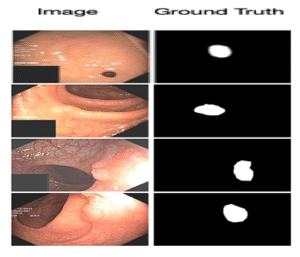 11
11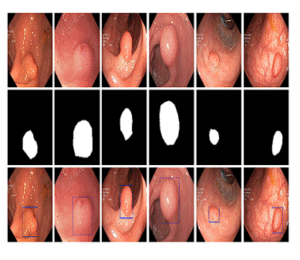 12
12 13
13 14
14 15
15KvasirCapsule-SEG
Segmentation dataset supporting lightweight real-time models such as NanoNet for capsule and colonoscopy workflows.
CapsuleEndoscopySegmentation
 16
16