Medical Image Segmentation Architectures

PVTFormer
[CT Liver Segmentation Via PVT-based Encoding and Refined Decoding] (ISBI 2024)]
PVTFormer is highly effective for healthy liver segmentation, with potential applications in other medical imaging areas. It represents a significant advancement in medical image segmentation, offering a robust solution for accurate diagnosis and treatment planning.
Github: https://github.com/DebeshJha/PVTFormer
Publication: https://arxiv.org/pdf/2401.09630
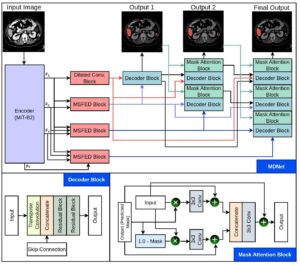
MDNet [MDNet: Multi-Decoder Network for Abdominal CT Organs Segmentation] [2024]
[MDNet: Multi-Decoder Network for Abdominal CT Organs Segmentation] [2024]
A MiT-B2 encoder extracts feature maps at four different levels (F1, F2, F3, and F4) from the input image. Each encoder network is linked to a specific part of the decoder through a multi-scale feature enhancement dilated block, increasing the network depth to generate three distinct segmentation masks. Additionally, the decoders are interconnected so that the output features from preceding decoders are utilized in the subsequent ones to refine the segmentation. Furthermore, the predicted masks from earlier decoders are incorporated into later stages to enhance feature map refinement. This approach ensures spatial attention across foreground and background regions, ultimately improving the final segmentation results.
Github: https://github.com/DebeshJha/MDNet
Publication: https://arxiv.org/pdf/2405.06166

TransNetR
[TransNetR: Transformer-based Residual Network for Polyp Segmentation with Multi-Center Out-of-Distribution Testing (MIDL 2023)]
TransNetR is an encoder decoder network which can be used for efficient biomedical image segmentation for both in-distribution and out-of-distribution datasets
Github: https://github.com/DebeshJha/TransNetR
Publication: https://arxiv.org/pdf/2303.07428

TransRUPNet
[TransRUPNet for Improved Out-of-Distribution Generalization in Polyp Segmentation]
We propose TransRUPNet, a real-time deep learning model using Transformers and residual upsampling for colorectal polyp segmentation. It features an encoder-decoder structure with upsampling blocks, achieving 47.07 FPS and a 0.7786 Dice score. Tested on OOD datasets, it outperforms existing methods, ensuring high accuracy and real-time feedback.
Github: https://github.com/DebeshJha/TransRUPNet
Publication: https://arxiv.org/pdf/2306.02176

DoubleUNet
[DoubleU-Net: A Deep Convolutional NeuralNetwork for Medical Image Segmentation]
DoubleU-Net starts with a VGG19 as encoder sub-network, which is followed by decoder sub-network. In the network, the input image is fed to the modified UNet(UNet1), which generates predicted masks (i.e., output1). We then multiply the input image and the produced masks (i.e., output1), which acts as an input for the second modified U-Net(UNet2) that produces another the generated mask (output2). Finally, we concatenate both the masks (output1 and output2) to get the final predicted mask (output).
Github: https://github.com/DebeshJha/2020-CBMS-DoubleU-Net
Publication: https://arxiv.org/pdf/2006.04868

ResUNet++
[ResUNet++: An Advanced Architecture for MedicalImage Segmentation]
The ResUNet++ architecture is based on the Deep Residual U-Net (ResUNet), which is an architecture that uses the strength of deep residual learning and U-Net. The proposed ResUNet++ architecture takes advantage of the residual blocks, the squeeze and excitation block, ASPP, and the attention block
Github: https://github.com/DebeshJha/ResUNetPlusPlus
Publication: https://arxiv.org/pdf/1911.07067

ResUNet++ + CRF + TTA
[A Comprehensive Study on Colorectal Polyp Segmentation With ResUNet++, Conditional Random Field and Test-Time Augmentation]
This is the extension of our previous version of the ResUNet++. In this paper, we describe how the ResUNet++ architecture can be extended by applying Conditional Random Field (CRF) and Test-Time Augmentation (TTA) to further improve its prediction performance on segmented polyps.
Github: https://github.com/DebeshJha/ResUNetPlusPlus-with-CRF-and-TTA
Publication: https://arxiv.org/pdf/1911.07067
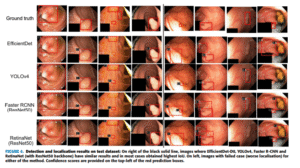
ColonSegNet [Real-Time Polyp Detection, Localization and Segmentation in Colonoscopy Using Deep Learning]
[ColonSegNet, a real-time deep learning model that outperforms existing methods in both accuracy and speed for polyp detection and segmentation using the Kvasir-SEG dataset.]
Computer-aided methods for detection, localization, and segmentation can enhance colonoscopy procedures, yet benchmarking these approaches remains challenging due to the growing variety of computer vision techniques applicable to polyp datasets. In this paper, we benchmark several state-of-the-art methods using the Kvasir-SEG dataset and demonstrate that our proposed ColonSegNet achieves the best trade-off between accuracy and real-time speed, with 0.8000 average precision and 180 FPS for detection, and a dice coefficient of 0.8206 with 182.38 FPS for segmentation. Our study highlights the critical role of standardized benchmarking in ensuring reproducibility and advancing reliable, real-time AI tools for clinical use.
Github: https://github.com/DebeshJha/
Publication: https://arxiv.org/pdf/1911.07067
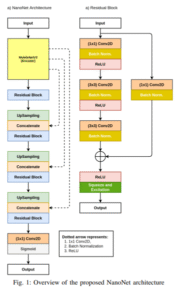
NanoNet
[NanoNet: Real-Time Polyp Segmentation in VideoCapsule Endoscopy and Colonoscopy]
This work introduces NanoNet, a lightweight deep learning architecture designed for real-time segmentation of video capsule endoscopy and colonoscopy images, achieving high accuracy with minimal computational cost. With only ~36,000 parameters, NanoNet outperforms more complex models in balancing speed, model size, and segmentation quality, making it suitable for integration into low-end clinical hardware.
Github: https://github.com/DebeshJha/
Publication: https://arxiv.org/pdf/2104.11138
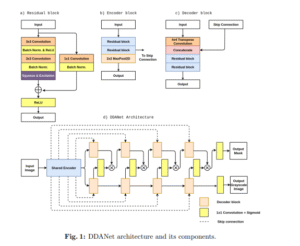
DDANet
[DDANet: Dual Decoder Attention Network for Automatic Polyp Segmentation]
This paper presents DDANet, a dual decoder attention network designed for accurate and efficient polyp segmentation in colonoscopy images. Trained on Kvasir-SEG and evaluated on an unseen dataset, DDANet demonstrates strong generalization with a dice coefficient of 0.7874 and precision of 0.8577, addressing challenges like variation and noise in polyp appearance.
Github: https://github.com/DebeshJha/
Publication: https://arxiv.org/pdf/2012.15245
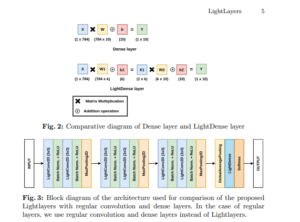
LightLayers
[LightLayers: Parameter Efficient Dense and Convolutional Layers for Image Classification]
This paper introduces LightLayers, a novel approach using matrix factorization to reduce the number of trainable parameters in deep neural networks, enabling faster training and lower computational demands. Tested on multiple benchmark datasets, LightLayers achieves competitive accuracy while significantly reducing model size, making deep learning more accessible to resource-constrained environments.
Github: https://github.com/DebeshJha/
Publication: https://arxiv.org/pdf/2101.02268
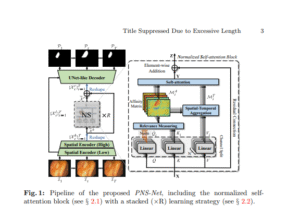
PNS-Net
[Progressively Normalized Self-Attention Network for Video Polyp Segmentation]
This paper presents PNS-Net, a real-time video polyp segmentation model based solely on normalized self-attention, overcoming CNN limitations by capturing global spatio-temporal information without post-processing. Achieving ~140 FPS and state-of-the-art performance on VPS benchmarks, PNS-Net proves effective through extensive evaluation of its progressive learning and attention strategies.
Github: https://github.com/DebeshJha/
Publication: https://arxiv.org/pdf/2105.08468
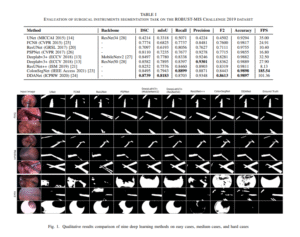
UNet
[U-Net Architecture for Surgical Image Segmentation (ROBUST-MIS-Challenge-dataset]This paper evaluates deep learning methods for automated segmentation of surgical instruments in minimally invasive surgery, a key step toward real-time tool tracking. The proposed DDANet achieves superior performance on the ROBUST-MIS 2019 dataset with a Dice coefficient of 0.8739, mIoU of 0.8183, and real-time speed of 101.36 FPS, making it suitable for clinical integration.
Github: https://github.com/DebeshJha/
Publication: https://arxiv.org/pdf/2107.02319
More information about the codes can be found at my GitHub webpage and publications can be found in Google Scholar