Medical AI Models Gallery
Browse a curated set of segmentation and vision architectures. Use the filters or search to quickly find what you need.
Results
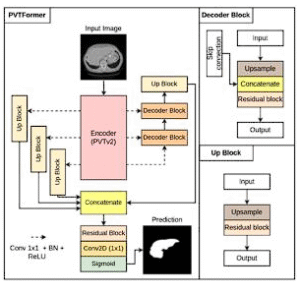
PVTFormer
TransformerISBI 2024CT Liver
PVT-based encoder with refined decoding for accurate, robust healthy-liver segmentation. Extensible to other modalities and tasks.
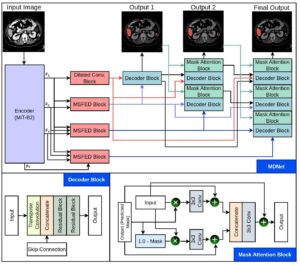
MDNet
Transformer EncoderAbdominal CT2024
MiT-B2 encoder with interlinked decoders and mask reuse to refine features, enforce spatial attention, and boost accuracy.
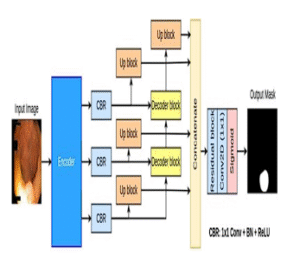
TransNetR
TransformerPolypMIDL 2023
Transformer-based residual network for robust polyp segmentation across in-distribution and out-of-distribution datasets.

TransRUPNet
TransformerReal-timePolyp
Encoder-decoder with residual upsampling blocks, 47.07 FPS and 0.7786 Dice, strong OOD generalization with real-time feedback.
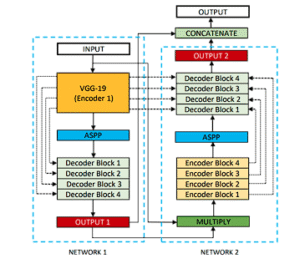
DoubleUNet
CNNTwo-Stage
VGG19-powered U-Net followed by a second U-Net; first mask multiplies input for refined second-stage segmentation.
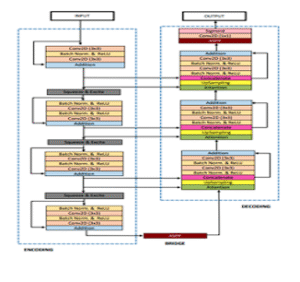
ResUNet++
CNNResidual
Residual U-Net enhanced with squeeze-and-excitation, ASPP, and attention blocks for stronger contextual feature learning.

ResUNet++ + CRF + TTA
CNNCRFTTA
Extends ResUNet++ with conditional random fields and test-time augmentation to further improve polyp segmentation quality.
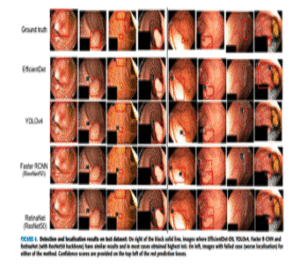
ColonSegNet
LightweightReal-timePolyp
Real-time model balancing accuracy and speed on Kvasir-SEG (~180 FPS, Dice ~0.8206), enabling reliable clinical feedback.
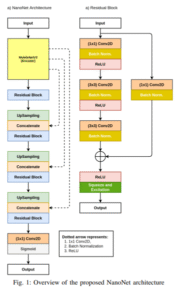
NanoNet
Lightweight~36k paramsReal-time
Ultra-compact architecture for real-time segmentation in video capsule endoscopy and colonoscopy with minimal compute.
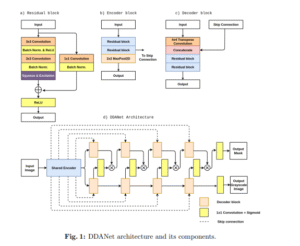
DDANet
CNNDual Decoder
Dual-decoder attention network trained on Kvasir-SEG, evaluated on unseen data with strong precision and Dice scores.
LightLayers
Machine LearningParam-Efficient
Matrix-factorized dense/conv layers reduce trainable parameters and speed up training while maintaining competitive accuracy.

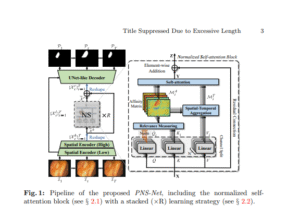
PNS-Net
TransformerVideoReal-time
Progressively normalized self-attention for video polyp segmentation, ~140 FPS and state-of-the-art VPS performance.
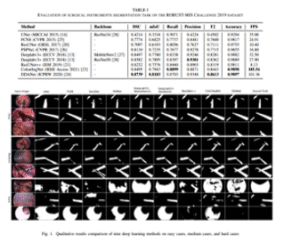
U-Net (ROBUST-MIS)
CNNSurgicalReal-time
Automated surgical instrument segmentation on ROBUST-MIS 2019 with high Dice and real-time throughput.